Research topics
Our research is focused on natural products as essential compounds for medical and biological applications. Based on the enormous value of natural products for the treatment of infections and tumors we focus on natural products that are active in these areas in particular. Synthetic access to natural products is always the most important aspect in our research. In this context, new strategies to novel structural motives have to be developed and applied to total synthesis. The vinylogous Mukaiyama aldol reaction is such a transformation on which we have focused during the past decades.
Since synthesis always requires a structure as target, we additionally developed a statistical tool to predict natural products based on the amino acid sequence of enzymes that are responsible for the biosynthesis. The so-called Hidden-Markov-Modell now allows structure prediction with good confidence.
An additional aspect of natural products chemistry stems for the combination of structure prediction through „gene analysis“ and the fact that principally every structure can be synthesized. The isolated natural products are not always identical with the structural prediction derived from the biosynthetically analysis. This can be seen as part of an evolutionary process. Through synthesis of these genetically encoded but not isolated natural products, we can trace back the evolution of natural products and learn how nature optimizes the desired biological activity.
Total synthesis
Our synthetic targets are derived from microorganisms, plants or marine sources. Below are some natural shown of which we have already completed their total synthesis.
- Aetheramid A, Chivosazol, Chlorotonil A, Corallopyronin A, Haprolid, Kulkenon, Lipomycin, Omphadiol, Tedanolid
Vinylogous Mukaiyama aldol reactions (VMAR)
We have developed diastereoselective as well as enantioselective vinylogous Mukaiyama aldol reactions on different substrates and substitution patterns.
-
Diastereoselective VMAR
Tris(pentafluorophenyl)borane-hydrate (TPPB•H2O) works best in our hands with excellent Felkin selectivity.
![]()
![]()
![]()
-
Enantioselective VMAR
Enantioselective VMARs mostly use different chiral Lewis acids and, basically all possible methyl group patterns can be obtained specifically.
![]()
![]()
![]()
Evolutionary analysis
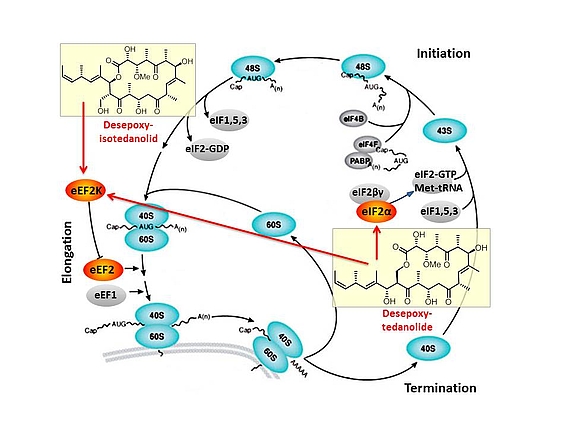
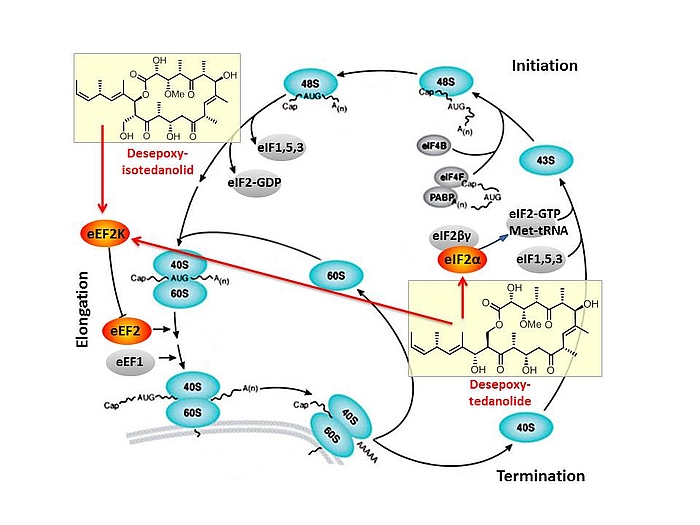
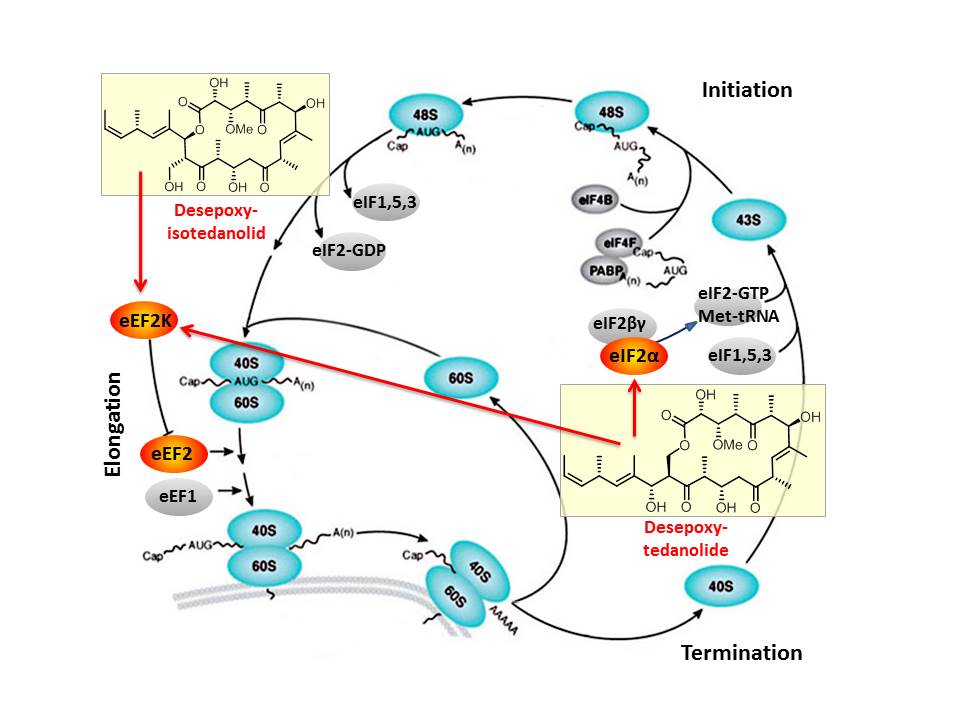
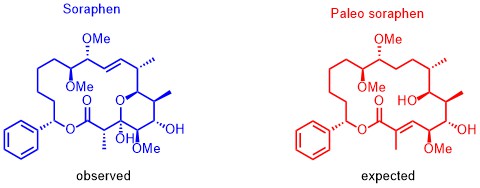


Based on our capabilities to make structure predictions form analyzing keto-reductases and being able to generate all polyketides at will, we started a program to synthesize natural products based on the predicted structure and not on the observed structure. With this, we try to get insight into how microorganisms optimize the biological activity of natural products for their needs.
In this context, we synthesized and evaluated the biological properties of the iso-tedanolide and paleo-soraphen. By generating a different lactone, tedanolide is able to bind to two targets in the translation process whereas iso-tedanolide binds only to one cellular target. Thus the biosynthetically not expected transesterification lead to a more potent cytotoxic compound.
The same research idea was applied to soraphen. By analyzing the biosynthesis clusters, we realized that the expected structure differs significantly from the isolated structure. By synthesizing the putative product and analyzing their biological activities we observed a completely different biological behavior. The natural soraphen is a bacterial acetyl-CoA carboxylase inhibitor whereas paleo-soraphen parallels the activity of a topoisomerase inhibitor. This indicates that microorganisms may also change the addressed target through significant structural changes of the natural products. Again, we are expecting even more insight in how microorganisms use natural products to address cellular targets and the power of synthesis is the essential tool to answer these biological questions.
Statistical structure prediction
Our structure prediction works for Type I Polyketides. Through analyzing keto-reduktases one can predict the configuration at secondary alcohols as well as at adjacent methyl branches.
Hidden-Markov-Modell
The required Hidden-Markov-Modell together with the required data set is available online. Just insert your keto-reductase sequence and run the programme. The output generates the Fischer nomenclature descriptors D/L as well as the absolute figure as an indication of the confidence of the prediction.
Ideally, the input of the sequence starts with positions TGGT or a corresponding sequence and ends with AWG or correspondingly.
Contact


30167 Hannover













